Скачать с ютуб Mutant-making: protein expression construct terms and site-directed mutagenesis в хорошем качестве
Скачать бесплатно и смотреть ютуб-видео без блокировок Mutant-making: protein expression construct terms and site-directed mutagenesis в качестве 4к (2к / 1080p)
У нас вы можете посмотреть бесплатно Mutant-making: protein expression construct terms and site-directed mutagenesis или скачать в максимальном доступном качестве, которое было загружено на ютуб. Для скачивания выберите вариант из формы ниже:
Загрузить музыку / рингтон Mutant-making: protein expression construct terms and site-directed mutagenesis в формате MP3:
Если кнопки скачивания не
загрузились
НАЖМИТЕ ЗДЕСЬ или обновите страницу
Если возникают проблемы со скачиванием, пожалуйста напишите в поддержку по адресу внизу
страницы.
Спасибо за использование сервиса savevideohd.ru
Mutant-making: protein expression construct terms and site-directed mutagenesis
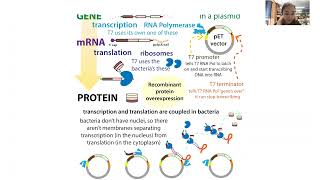
With site-directed mutagenesis, I can make specific mutations in proteins by changing their DNA instructions. This lets me see what different parts of proteins do. And I’ve done it A LOT over the past 5 years. As I’m writing up my thesis and paper, I keep coming across a lot of “construct” nomenclature and it really hit home how important site-directed mutagenesis has been to my project! And how confusing some of the terms first seemed, so here we go. ⠀ blog form with full text & figs: https://bit.ly/mutagenesisconstructs It’s a lot easier to edit DNA when it’s in a small circular piece of DNA called a plasmid inside a test tube than when it’s in a huge coiled-up chromosome inside a cell. So, if we want to study a protein, we can stick the genetic instructions for the protein into a circular piece of DNA called a plasmid vector. This is called molecular cloning and I discussed it in depth here: http://bit.ly/molecularcloningguide ⠀ ⠀ We can then stick that into cells (often bacteria) to get them to make more of the plasmid containing our inserted genetic instructions, and we can stick the plasmid into expression cells to make the corresponding protein. We call this RECOMBINANT protein production. ⠀ ⠀ Once the DNA is in the plasmid, we can use site-directed mutagenesis techniques like SLIC mutagenesis and QuikChange mutagenesis to introduce specific DNA letter changes into the genetic instructions for the protein. And this can lead to specific protein changes. ⠀ ⠀ If you’re wondering why we might want to do this, think of a spoon versus a knife. Structure (bowliness vs. long flat jaggedness) and function (scooping vs slicing) are intimately connected. ⠀ ⠀ The same holds true for biochemical “utensils” like proteins, and we can exploit this relationship to learn about function from structure and structure from function by making specific changes to specific parts using site-directed mutagenesis, then testing to see if those changes affected the protein’s activity. ⠀ ⠀ A protein’s structure comes ultimately from its genetic instructions: the sequence of DNA letters that get copied into RNA letters determines the sequence of proteins letters (amino acids) in a protein. And because those different amino acids have different properties (size, charge, water-likingness), they affect how the protein folds (structure) & how it acts (function). So if we change the gene, we can change the protein and potentially its functioning.⠀ ⠀ Potentially because it all depends on where you mutate and how drastic the change is. If, for example, you pierced a small hole in the cup of a spoon and the blade of a knife you probably wouldn’t be able to tell anything had changed if you tested out the knife. But if you tried slurping some soup with that spoon you’d probably dribble down your front….⠀ ⠀ With site-directed mutagenesis you can do things like make⠀ - substitutions (change some DNA lteters)⠀ - these can be drastic (such as changing a negatively-charged aspartate to a positively-charged arginine) or “conservative” (such as shifting around the atoms of leucine to get isoleucine which acts pretty darn similarly)⠀ - insertions (add some DNA lettttters)⠀ - deletions (remove some DNA ltrs)⠀ - when we shorten the ends we typically refer to these changes as “truncations.” If you cut off part of the “starting end” of the protein we call it an N-terminal truncation and if you cut off part of the “ending end” we call it a C-terminal truncation⠀ ⠀ We refer to the original, “non-mutated” form as “wild-type” and the changed versions as “mutants”⠀ ⠀ Some notations we use to describe what we’ve done to it:⠀ - For substitutions, we put the original letter first & what it’s changed to afterwards. So, say you changed the 91st amino acid from a glycine (G) to an alanine (A). You could write this as G91A. And if you changed the 215th amino acid from A to G you’d write A215G⠀ - to indicate taking out stretches of amino acids, we use the delta sign, Δ. So if we removed amino acids 80-100, we would write Δ80-100. ⠀ How do we do it? I like SLICE (Site & Ligation Independent Cloning) but another common way is QuikChange. More details in the blog version