Скачать с ютуб Dorothee Kern (Brandeis, HHMI) 1: Visualizing Protein Dynamics в хорошем качестве
Скачать бесплатно и смотреть ютуб-видео без блокировок Dorothee Kern (Brandeis, HHMI) 1: Visualizing Protein Dynamics в качестве 4к (2к / 1080p)
У нас вы можете посмотреть бесплатно Dorothee Kern (Brandeis, HHMI) 1: Visualizing Protein Dynamics или скачать в максимальном доступном качестве, которое было загружено на ютуб. Для скачивания выберите вариант из формы ниже:
Загрузить музыку / рингтон Dorothee Kern (Brandeis, HHMI) 1: Visualizing Protein Dynamics в формате MP3:
Если кнопки скачивания не
загрузились
НАЖМИТЕ ЗДЕСЬ или обновите страницу
Если возникают проблемы со скачиванием, пожалуйста напишите в поддержку по адресу внизу
страницы.
Спасибо за использование сервиса savevideohd.ru
Dorothee Kern (Brandeis, HHMI) 1: Visualizing Protein Dynamics
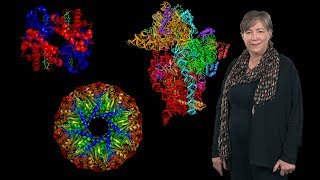
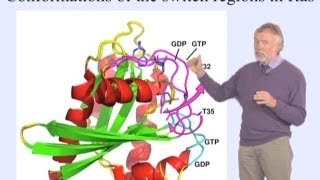
https://www.ibiology.org/biophysics/p... Dorothee Kern explains how visualizing protein dynamics (i.e. watching proteins in action) allows us to better understand protein function and optimize drug design. Talk Overview: Proteins such as signaling molecules, catalytic enzymes and membrane transporters, are not static but are in a state of constant motion for function. In her first talk, Dorothee Kern explains how she uses techniques such as nuclear magnetic resonance (NMR), X ray crystallography, single molecule FRET and computational simulations to visualize rapid protein dynamics. Kern walks us through the basics of these techniques and explains how they can provide structural and energetic information about a protein and how it functions in the cell. Gleevec (Imatinib) is a very effective drug for treating chronic myelogenous leukemia. It acts by binding to and inhibiting the signaling molecule Abl kinase with extreme specificity. Abl kinase is very closely related to Src kinase, and, in fact, the drug binding pocket for Gleevec is almost identical between the two proteins. Interestingly, however, Gleevec binds to Abl 3000 fold more tightly than it does to Src. Why? In her second talk, Kern answers this question. Her lab used NMR, and the techniques she described in Part 1, to show that the protein dynamics of Abl and Src are dramatically different when Gleevec is bound. In a clever experiment in which they synthesized proteins that were likely evolutionary ancestors of Src and Abl, Kern’s lab was able to show how changes in amino acids throughout the kinases determined the differential binding affinity of Gleevec for Abl kinase over Src kinase. Experiments such as these demonstrate the importance of understanding protein dynamics at the atomic level of the whole protein, not just the drug binding site, when designing new drugs. Speaker Biography: Dorothee Kern is a Professor of Biochemistry at Brandeis University and an Investigator of the Howard Hughes Medical Institute. Kern strives to understand how proteins function by understanding how they move. She uses biophysical analytical methods, in particular nuclear magnetic resonance (NMR), to analyze protein dynamics. Using NMR, she has followed the high speed motion of enzymes during catalysis and provided unexpected insights into how conformational change can speed up biological reactions. She has recently expanded the concept of protein motion to the evolution of proteins over billions of years by reconstructing evolutionary trajectories in vitro. Kern pursues a new vision of protein dynamics and allosteric networks at the heart of improved drug design with the mission of exploiting protein dynamics for expanding therapeutics. Kern received her BS, MS and PhD in biochemistry from Martin Luther University in Halle, Germany. Before moving to California for her post-doc at UC Berkeley and the Lawrence Berkeley National Laboratory, Kern played on the German National Basketball team for ten years. In 1999, Kern joined the faculty of Brandeis University and transferred her leadership skills from the basketball court to the lab. Kern’s insights into protein dynamics and function have been recognized with the Pfizer Award in Enzyme Chemistry and the Dayhoff and National Lecture Awards from the Biophysical Society. Learn more about Kern’s research here: http://www.bio.brandeis.edu/kernlab/ and http://www.hhmi.org/scientists/doroth...