Скачать с ютуб prolfqua: A comprehensive R package for protein differential expression analysis в хорошем качестве
Скачать бесплатно и смотреть ютуб-видео без блокировок prolfqua: A comprehensive R package for protein differential expression analysis в качестве 4к (2к / 1080p)
У нас вы можете посмотреть бесплатно prolfqua: A comprehensive R package for protein differential expression analysis или скачать в максимальном доступном качестве, которое было загружено на ютуб. Для скачивания выберите вариант из формы ниже:
Загрузить музыку / рингтон prolfqua: A comprehensive R package for protein differential expression analysis в формате MP3:
Если кнопки скачивания не
загрузились
НАЖМИТЕ ЗДЕСЬ или обновите страницу
Если возникают проблемы со скачиванием, пожалуйста напишите в поддержку по адресу внизу
страницы.
Спасибо за использование сервиса savevideohd.ru
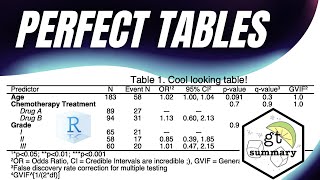
prolfqua: A comprehensive R package for protein differential expression analysis
What you will discover: How prolfqua can be used for analysis of protein expression in data generated from mass spectrometry. You are: Interested in measuring levels of proteins in biological samples or in analyzing data generated from mass spectrometry. Speaker: Witold E. Wolski, Data Scientist at Proteome Informatics led by Christian Panse. Click to access specific sections of the talk: - 01:11 Protein quantification with mass spectrometry - 02:30 Protein differential expression analysis - Challenges - 04:56 Functionality and Implementation - 07:02 Usage example - 09:25 Performance/Benchmarking - 12:19 Outlook Reference papers and resource Wolski et al., prolfqua: A Comprehensive R-Package for Proteomics Differential Expression Analysis. Journal of proteome research 2023. DOI: 10.1021/acs.jproteome.2c00441 Any questions about this talk? Contact Witold at witold.wolski[at]fgcz.uzh.ch More about the speaker: Witold E. Wolski obtained his doctorate in computational chemistry from the Free University Berlin, Germany, under the supervision of Knut Reinert, after which he undertook postdoctoral research at the School of Mathematics and Statistics at the University of Newcastle, UK. He worked for five years as a software engineer at Bruker Daltonics in Bremen, Germany, where he developed software for mass spectrometry. Then, he moved to Switzerland to provide bioinformatics support in the laboratory of Ruedi Aebersold. Witold has contributed to computational mass spectrometry and proteomics for over two decades and now works at the SIB Group led by Christian Panse at the Functional Genomics Center Zurich. His research interests include mathematical and statistical modeling of mass spectrometric proteomics and metabolomics data and data analysis automation. Read the news on our website: https://www.sib.swiss/in-silico-talks...