Скачать с ютуб Understanding Quantitative Trait Loci with the help of tomatoes в хорошем качестве
Скачать бесплатно и смотреть ютуб-видео без блокировок Understanding Quantitative Trait Loci with the help of tomatoes в качестве 4к (2к / 1080p)
У нас вы можете посмотреть бесплатно Understanding Quantitative Trait Loci with the help of tomatoes или скачать в максимальном доступном качестве, которое было загружено на ютуб. Для скачивания выберите вариант из формы ниже:
Загрузить музыку / рингтон Understanding Quantitative Trait Loci with the help of tomatoes в формате MP3:
Если кнопки скачивания не
загрузились
НАЖМИТЕ ЗДЕСЬ или обновите страницу
Если возникают проблемы со скачиванием, пожалуйста напишите в поддержку по адресу внизу
страницы.
Спасибо за использование сервиса savevideohd.ru
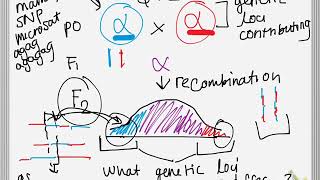
Understanding Quantitative Trait Loci with the help of tomatoes
A review of: Resolution of quantitative traits into Mendelian factors by using a complete linkage map of restriction fragment length polymorphisms. Paterson AH, Lander ES, Hewitt JD, Peterson S, Lincoln SE, Tanksley SD. Nature. 1988 Oct 20;335(6192):721-6. doi: 10.1038/335721a0. Twitter: / genetics_stuff OTHER VIDEOS YOU MIGHT LIKE: • Proving the linear structure of genes: Benzer shoots and scores a goal (Benzer, 1959) - • Proving the linear structure of genes... • Plants show phenotype is influenced by genotype AND environment (Clausen, Keck & Hiesey, 1940) - • Plants show phenotype is influenced b... • Codons don’t overlap in the shagadelic genetic code, baby. Yeah! (Brenner, 1957) - • Codons don’t overlap in the shagadeli... Genetic traits can be separated into two broad categories; simple traits which have discrete phenotypes and complex traits which have continuous phenotypes. Complex traits are controlled by a finite number of genes; each with a discrete influence on the overall phenotype. However, the number of genes is so large that the impact of any one gene is overshadowed by environmental variation to give the illusion of continuity. To make things more difficult, the nature of independent assortment makes it extremely difficult to study any complex trait gene in isolation. In 1988, Paterson and Lander sought to overcome the difficulties associated with studying complex traits in their study of tomato fruit mass, pH, and soluble solid concentration. Their approach involved locating quantitative trait loci (QTLs) by their association with DNA polymorphisms associated with the recognition sites of common restriction enzymes – known as restriction fragment length polymorphisms (RFLPs). Paterson and Lander’s RFLP map contained 70 markers with an average density of 14.3 cM across all twelve chromosomes. By studying the offspring of Lycopersicon esculentum and Lycopersicon chmielewski, parents with vastly different phenotypes, the researchers were able to genotype and phenotype plants to identify the position and effect of fourteen QTLs spread across the three traits. The success of the Paterson and Lander encouraged other scientists to follow suit, creating QTL maps for many of the world’s most important crops. SNP markers have since made it possible to genotype changes in a single nucleotide and therefore at a much higher resolution, and QTL analysis has now been applied in higher order mammals to understand the genetic factors which underpin human disease. Creator: John Saxon References: Anderson J., Stack R., Liu S., Waldron B., Fjeld A., Coyne C., Moreno-Sevilla B., Fetch J., Song Q., Cregan P., Frohberg R., 2001 DNA markers for Fusarium head blight resistance QTLs in two wheat populations. Theoretical and Applied Genetics 102: 1164-1168. Bernardo R., 2008 Molecular markers and selection for complex traits in plants: Learning from the last 20 years. Crop Science 48: 1649-1664. Frayling T., 2014 Genome-wide association studies: the good, the bad and the ugly. Clinical Medicine 14: 428-431. Kumar S., Banks T., Cloutier S., 2012 SNP Discovery through next-generation sequencing and its applications. International Journal of Plant Genomics 2012: 1-15. Lander E., Green P., 1987 Construction of multilocus genetic linkage maps in humans. Proceedings of the National Academy of Sciences 84: 2363-2367. Paterson A., Lander E., Hewitt J., Peterson S., Lincoln S., Tanksley S., 1988 Resolution of quantitative traits into Mendelian factors by using a complete linkage map of restriction fragment length polymorphisms. Nature 335: 721-726. Singh R., Singh S., Tiwari D., Srivastava S., Singh S., Sharma M., Singh R., Mohapatra T., Singh N., 2012 Genetic mapping and QTL analysis for sugar yield-related traits in sugarcane. Euphytica 191: 333-353. Smýkal P., K. Varshney R., K. Singh V., Coyne C., Domoney C., Kejnovský E., Warkentin T., 2016 From Mendel’s discovery on pea to today’s plant genetics and breeding. Theoretical and Applied Genetics 129: 2267-2280. Xu S., 2012 Interval Mapping. In: Xu S Principles of Statistical Genomics, Springer, New York, pp. 109-129.